Home
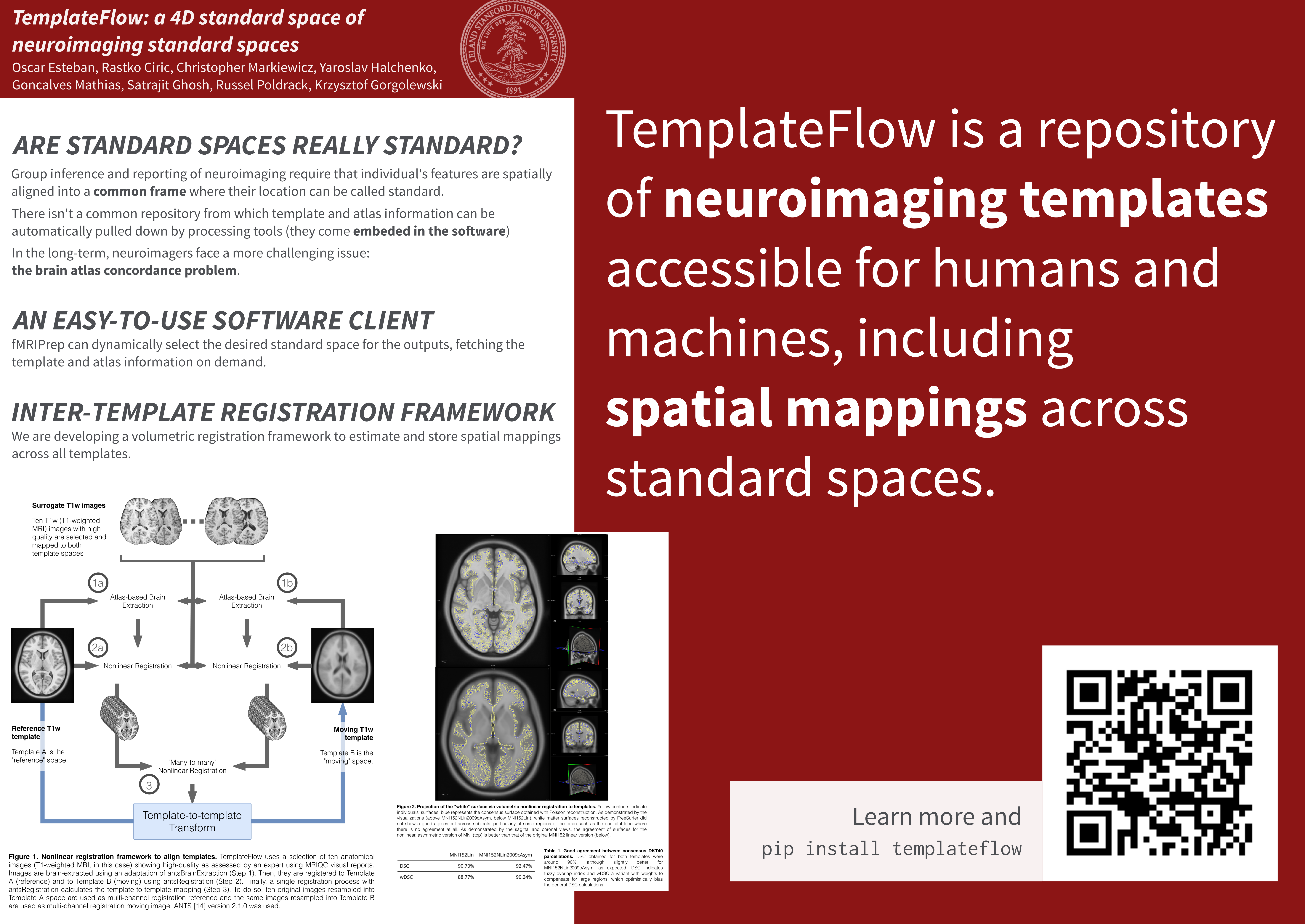
Group inference and reporting of neuroimaging studies require that individual's features are spatially aligned into a common frame where their location can be called standard (Brett et al., 2002). To that end, a multiplicity of brain templates with anatomical annotations (i.e. atlases) have been published (Dickie et al., 2017). However, a centralized resource that allows programmatic access to templates is lacking. TemplateFlow is a modular, version-controlled resource that allows researchers to use templates "off-the-shelf" and share new ones.
 |
|---|
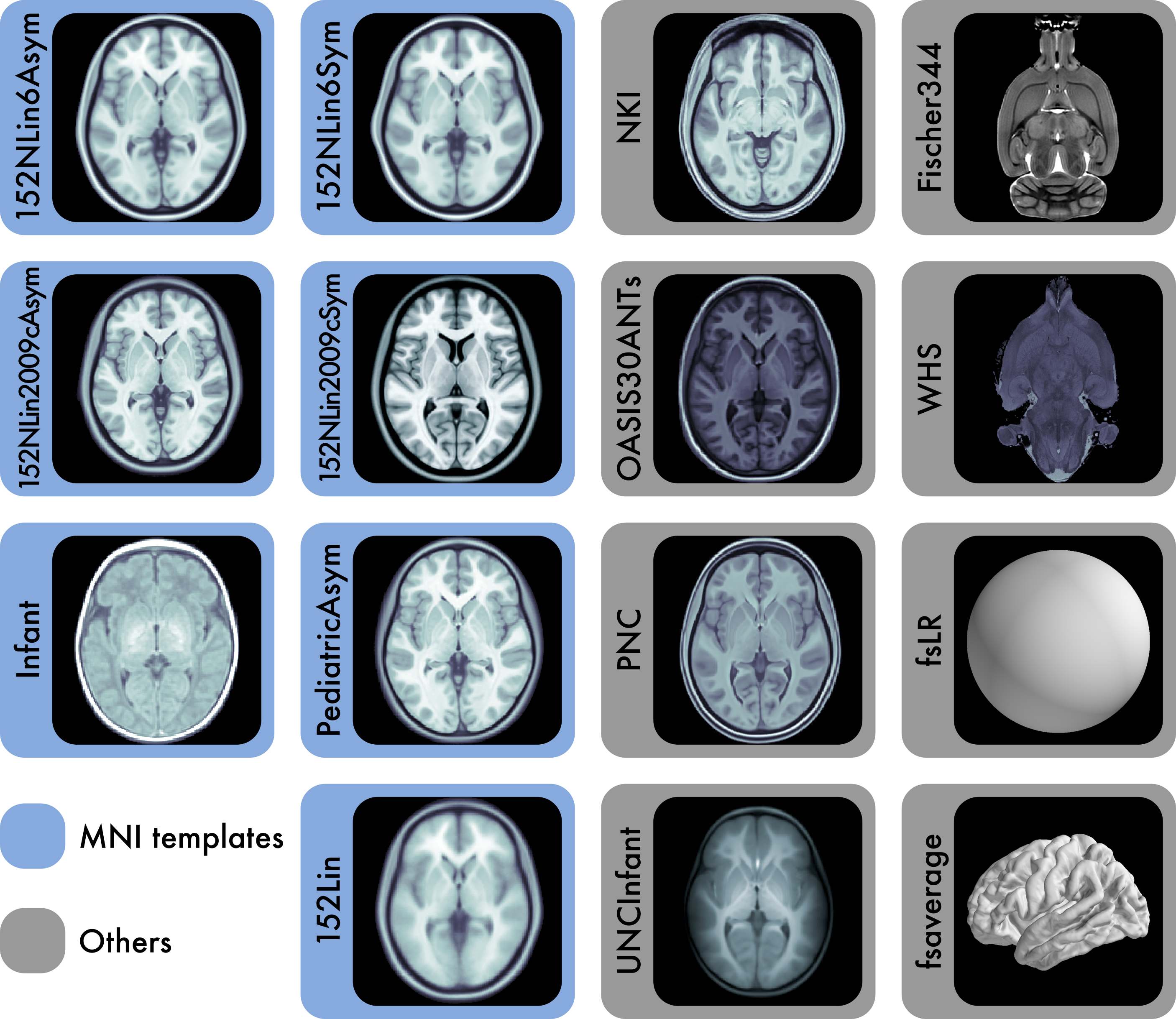
The 7 templates highlighted in blue are constituents of the Montreal Neurological Institute (MNI) portfolio. The Waxholm space (WHS) and Fischer344 templates provide references for rat neuroimaging. fsaverage and fsLR are surface templates; the remaining templates are volumetric. Each template is distributed with atlas labels, segmentations, and metadata files. The 15 templates displayed here are only a small fraction of those created as stereotaxic references for the neuroimaging community. |
Please head over to our TemplateFlow Archive browser.
Bird's view over TemplateFlow¶
 |
|---|
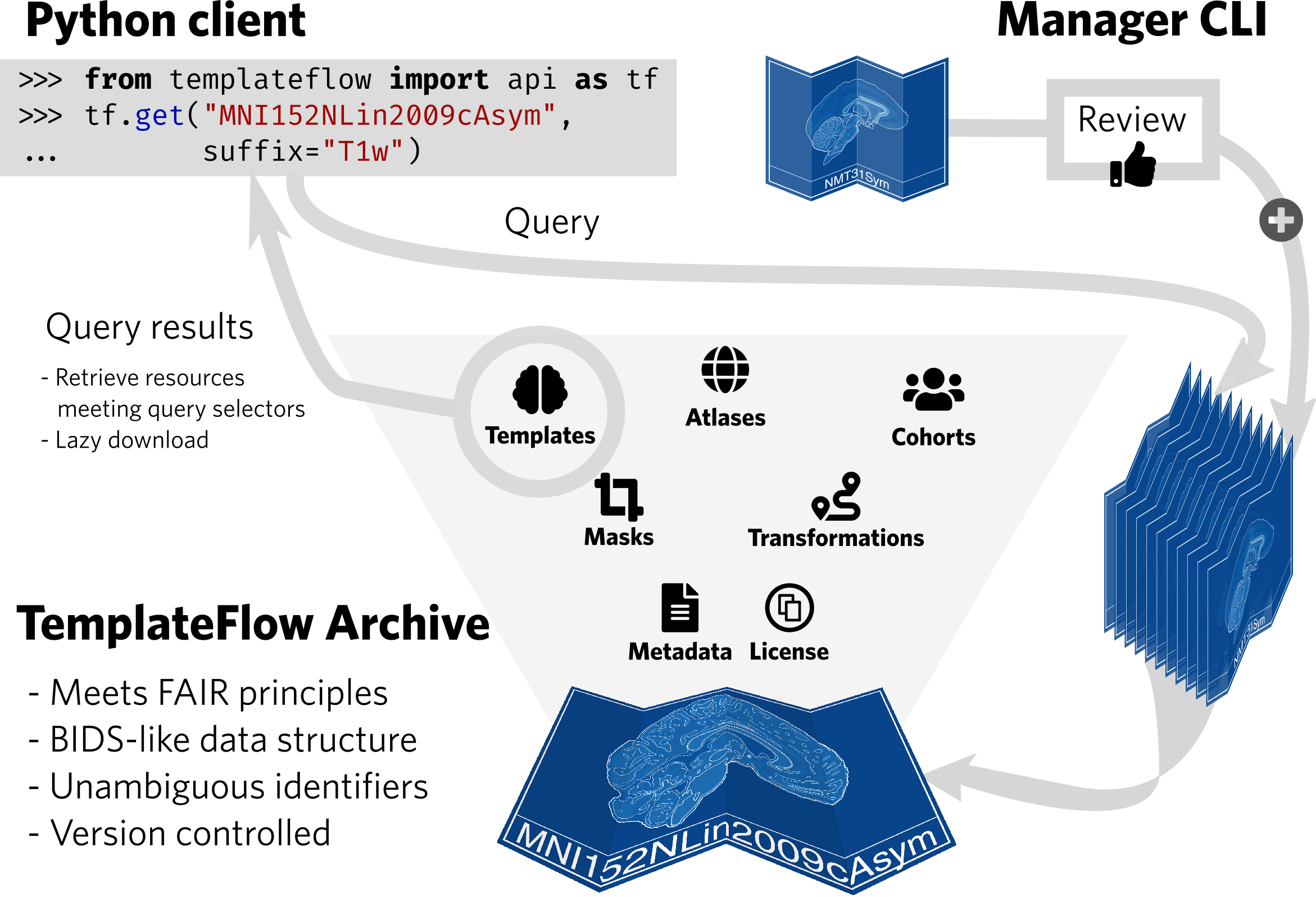
| TemplateFlow implements the FAIR guiding principles. The TemplateFlow Archive can be accessed at a “low” level with DataLad, or at a “high” level with a Python client. New resources can be added through the TemplateFlow Manager Command Line Interface, which initiates a peer-review process before acceptance in the Archive. |